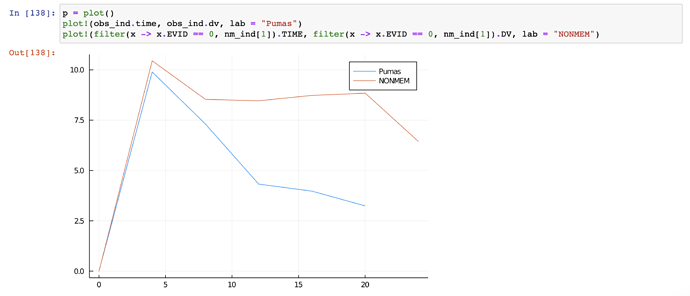
Hello there! I am new to Pumas and have trying to validate some of my previous results from NONMEM with Pumas. I have read through the docs nearly 10 times, and I got some of my other models to work, but for some reason, one of my drugs is producing results that are consistently too low. I cannot seem to identify the structural difference between the models that could be causing this, and I have tested and ensured that the error model is not the cause.
Here is the NONMEM model:
$PROBLEM Simulation Levofloxacin PK
$INPUT ID TIME EVID AMT DV TAD DROP SEX AGE WT HT WAZW WHZ HAZW BAZW COUNTRY ; Dropped TAD HIV ... Country
$DATA ../pop_sample1000_test.csv IGNORE=@
; AMT in mg
; DV in ug/mL or mg/L
; TIME in hours
$SUBROUTINE ADVAN6 TOL=5
$MODEL NCOMP=4
COMP=(DEPOT DEFDOS)
COMP=(CENTRAL DEFOBS)
COMP=(PERIPH)
COMP=(AUC)
$PK
BSIZE1 = WT/12 ; This needed to be changed to 12 kg based on the study. see footnote a in table 2.
EXPO = 0.75
CLHIV = 1 + 0*THETA(8)
; AGE EFFECT
GAM = THETA(11)
PMAGE50 = THETA(10)
MAT = AGE**GAM/(PMAGE50**GAM + AGE**GAM) ; AGE in months
; START PK
TVCL = THETA(1)*CLHIV*MAT*BSIZE1**EXPO ; added maturation on CL, removed HAZ.
CL = TVCL*EXP(ETA(1)) ; CL, L/h [systemic clearance]
TVV = THETA(2)
V = TVV*BSIZE1*EXP(ETA(2)) ; V, L [central volume]
TVKA = THETA(3) ; KA, 1/h [absorption rate]
KA = TVKA*EXP(ETA(3))
TVVP = THETA(6)
VP = TVVP*BSIZE1*EXP(ETA(4))
TVQ = THETA(7)
Q = TVQ*BSIZE1**EXPO*EXP(ETA(5))
TVF1 = 1
F1 = TVF1*EXP(ETA(6))
K20 = CL/V
K23 = Q/V
K32 = Q/VP
ALAG1 = THETA(9)*EXP(ETA(7))
$DES
DADT(1) = -KA*A(1)
DADT(2) = KA*A(1)-K20*A(2)-K23*A(2)+K32*A(3)
DADT(3) = K23*A(2)-K32*A(3)
DADT(4) = A(2)/V
$ERROR
REP = IREP
AUC = A(4)
; Proportional Residual Error model
CP = A(2)/V
IPRED = CP
PROP = THETA(4)
ADD = THETA(5)
W = SQRT(PROP**2*IPRED**2+ADD**2)
IRES = DV - IPRED
IWRES = IRES/W
Y = IPRED + W*EPS(1)
$THETA ; you need to change CL, V, Prop, Add, VP, Q ...
4.7 ; 1 CL,L/h [systemic clearance]
19.2 ; 2 Vc,L [central volume]
1.61 ; 3 Ka
0.116 ; 4 PROP
0.0160 ; 5 ADD
2.40 ; 6 VP
0.796 ; 7 Q
-0.159 ; 8 HIV on CL, study found 15.9% decrease in CL
0.242 ; 9 ALAG
10.6 ; 10 PMAGE50
3.39 ; 11 GAMMA
$OMEGA
0.023 ; CL IIV these need to be converted. 15% CV = .15^2 =
0 FIX ; V IIV
0.420 ; KA IOV ... we might want to consider inputting the BOV but for now it is fine.
0 FIX ; VP
0 FIX ; Q
0.048 ; F1
1.69 ; ALAG IOV
$SIGMA 1 FIX ; RV (use 20% CV as init est)
$SIM ONLYSIM NSUB=100 (12345)
$TABLE NOPRINT ONEHEADER FILE=simtab ID TIME EVID AMT TAD DV IPRED CP WT AUC REP WT AGE
And here is the Pumas model;
model = @model begin
@param begin
tvcl ∈ RealDomain(lower = 0) ; 1
tvv ∈ RealDomain(lower = 0) ; 2
tvka ∈ RealDomain(lower = 0) ; 3
σ_prop ∈ RealDomain(lower = 0) ; 4
σ_add ∈ RealDomain(lower = 0) ; 5
tvvp ∈ RealDomain(lower = 0) ; 6
tvq ∈ RealDomain(lower = 0) ; 7
clhiv ∈ RealDomain(lower = 0) ; 8
alag ∈ RealDomain(lower = 0) ; 9
pmage50 ∈ RealDomain(lower = 0) ; 10
γ ∈ RealDomain(lower = 0) ; 11
Ω ∈ PDiagDomain(7)
end
@random begin
η ~ MvNormal(Ω)
end
@covariates WT AGE
@pre begin
BSIZE1 = WT / 12
EXPO = 0.75
CLHIV = 1 + 0 * clhiv
VP = tvvp * BSIZE1 * exp(η[4])
Q = tvq * (BSIZE1^0.75) * exp(η[5])
MAT = (AGE^γ) / ((pmage50^γ) + (AGE^γ))
CL = tvcl * CLHIV * MAT * (BSIZE1^0.75) * exp(η[1])
V = tvv * BSIZE1 * exp(η[2])
KA = tvka * exp(η[3])
K20 = CL / V
K23 = Q / V
K32 = Q / VP
tvf1 = 1
bioav = (Depot = tvf1 * exp(η[6]),)
lags = (Depot = alag * exp(η[7]),)
end
@dynamics begin
Depot' = -KA*Depot
Central' = KA*Depot - Central*K20 - Central*K23 + K23*Peripheral
Peripheral' = K23*Central - K32*Peripheral
AUC' = Central / V
end
@vars begin
conc = Central / V
auc = AUC
end
@derived begin
cp = @. Central / V
dv ~ @. Normal(cp, sqrt(σ_add^2 + (cp*σ_prop)^2))
end
end
fixeffs = (
tvcl = 4.7,
tvv = 19.2,
tvka = 1.61,
σ_prop = 0.116,
σ_add = 0.0160,
tvvp = 2.4,
tvq = 0.796,
clhiv = -0.159,
alag = 0.242,
pmage50 = 10.6,
γ = 3.39,
Ω = Diagonal([0.023, 0, 0.420, 0, 0, 0.048, 1.69]),
)
obs = simobs(model, population, fixeffs, obstimes = 0:4:24) |> DataFrame
I apologize in advance if this post is too lengthy or otherwise out of the scope of what everyone can help with, but I appreciate any advice in advance, thank you!
Here is a histogram comparing the distributions of AUC between a single run of Pumas and a single run of NONMEM
And here is a histogram comparing the median AUC of a single Pumas run to the distribution of median AUC of 100 simulation runs in NONMEM
Thank you again!